Graduate School of Science and Engineering Applied Chemistry
- Course Outline
- Electrochemical Laboratory
- Laboratory of Physical Chemistry
- Polymer Chemistry Laboratory
- Laboratory of Inorganic Synthetic Chemistry
- Functional Organic Chemistry Laboratory
- Laboratory of Catalysis and Material Design
- Laboratory of Biofunctional Chemistry
- Powder Technology Laboratory
- Transport Phenomena Laboratory
- Biochemical Engineering Laboratory
- Separation and Detection Chemistry Laboratory (Analytical Chemistry Laboratory)
- Molecular Chemical Engineering
- Material System Laboratory
- Biosensing Laboratory
Biosensing Laboratory(Bioanalytical Chemistry Laboratory)
Development of diagnostic applications with fast, sensitive, and cost-efficient sensing manner Web site of the laboratory 【Currently not available】
Staff

HASHIMOTO Masahiko
[Professor]
| Acceptable course | |
|---|---|
| Master's degree course | ✓ |
| Doctoral degree course | |
Telephone : +81-774-65-6594
mahashim@mail.doshisha.ac.jp
Office : SC-324
Database of Researchers
Research Topics
The central theme of Dr. Hashimoto’s program is to create novel techniques for biomedical discovery and clinical diagnosis through the analysis of biological macromolecules including DNAs, RNAs, and proteins. Developing these techniques involves a wide range of research activities such as the creation of new molecular probes, novel assay and analytical methodologies, and device innovation. What is particularly attracting his research interest is that these techniques are being integrated into small operating platforms that allow to perform various diagnostic and prognostic applications of many human diseases (e.g., cancers). In order to establish such analytical platforms, his research covers a diverse range of subarea, such as photolithography-based microfabrication, photoluminescent probe development, construction of ultrasensitive detection instruments, and manipulation of micro- and nanofluids. Provided below is a brief description of a few of many projects in his group.
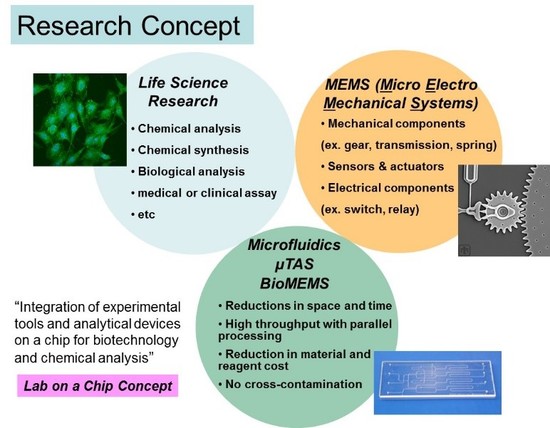
Research Contents
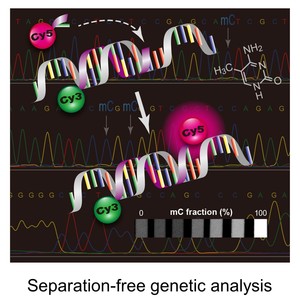
New assays for detecting biomarkers
Novel assays are being exploited to facilitate rapid reporting genetic and epigenetic biomarkers specific to a particular disease type. For instance, some of our projects are focused on designing and constructing a point-of-care molecular diagnostic test for cancer. We are using fluorescence monitoring approach to detect certain mutated DNAs that are sporadically present in normal DNA population when a patient experiences early carcinogenesis. Following isolation of genomic DNAs from human cells, the genomic DNAs are subjected to primary PCR amplification, followed by a ligation- or extension-based reaction, which allows formation of short DNA strands (i.e., ligation or extension products) only if the target mutation is present. The products contains a pair of fluorescent molecules at the ends of the strand. The dye-pair can undergo a fluorescence energy transfer (FRET) process that allow direct detection of the product without any separation step.
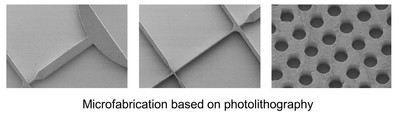
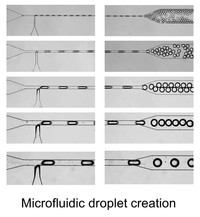
Instrumentation developments
One of our hardware developments is focused on designing, manufacturing, and evaluating microfluidic chips for molecular analyses. In addition to laser machining and focused ion beam milling, we are mainly using the so-called “soft lithography” technique to fabricate replicas of prerequisite polydimethylsiloxane (PDMS) chips that consist of fluidic channels, which range in size of >10 μm. We are currently able to fabricate complicated microstructures that differ in channel depth in a single chip with an application of a stepwise multiple processing of the photoresist coating followed by UV exposure for SU-8 mold fabrication. With the developed chips, we have established a unique method that allowed automatic preparation of monodisperse water-in-oil droplets at a rate of ca. 1 kHz without connecting external pumping devices for fluid control. This facile and convenient technique is spanning a range of droplet-based (bio)chemical applications in the relevant research field.
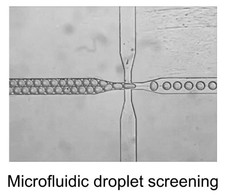

Another hardware development is a construction of high-performance capillary electrophoresis (CE) system for DNA sequencing and genotyping. While capillary array electrophoresis have greatly facilitated DNA sequencing, for a long time people have been concerned about how to integrate multiple steps involved in the DNA sequencing workflow such as sample preparation, PCR amplification, amplicon cleanup, and sequencing to take advantage of multiple array. Dr. Hashimoto and co-workers constructed a fully integrated sample preparation-PCR-cleanup-sequencing system. Because everything is conducted on-line in connected capillary tubes, it should be possible to upgrade this to a highly multiplexed format as well with minimal amounts of sample.

Single-molecule detection and imaging
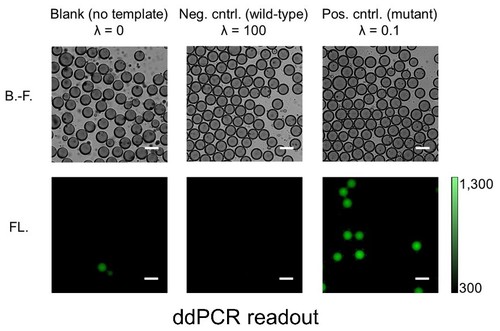
The capacity to selectively and sensitively detect single biomolecules such as DNA, RNA, and protein could bring enormous breakthroughs to chemistry, biology, and medicine. For example, the ability to disclose genetic status with the sensitivity at the single-molecule level will have considerable value in basic biomedical study, drug discovery, and clinical diagnosis. One of the most powerful technologies for nucleic acid test at the single-molecule level is PCR, owing to its dramatic amplification ability across several orders of magnitude that can produce millions of copies of a targeted DNA sequence even from a single copy. However, conventional PCR with low starting template concentration often demands critical requirements for the experimental parameters to obtain the maximal product yield while minimizing nonspecific amplicon. Later on, it was realized that reduction in reaction volume (e.g., 1 nL), which increased the effective concentration of the initial template, was very critical for successful single-molecule PCR. To further reduce the reaction volume without sacrificing amplification efficiency and throughput, aqueous droplets dispersed in a continuous oil phase were employed as discrete PCR reaction compartments. An analytical technique of highly parallel PCR within an enormous number of droplets followed by endpoint discrimination of fluorescence-positive (‘1’) and –negative (‘0’) droplets (i.e., digital sorting of fluorescence signature) is called droplet digital PCR (ddPCR). The ddPCR has recently attracted much attention since this new technique allowed various applications previously unattainable. These include copy number variation (CNV) analysis, quantification of DNA libraries for the Next-Generation Sequencing (NGS), differential gene expression analysis, rare mutation detection, and etc. While commercial ddPCR platforms are currently available, Dr. Hashimoto’s group has established a very facile, fast, and inexpensive ddPCR method using a degassed PDMS microfluidic device. This study does not only makes ddPCR technology more accessible to its potential users but also stimulate further development of other droplet-based applications.
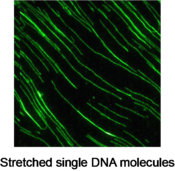
By using the total internal reflection fluorescence (TIRF) imaging system, Dr. Hashimoto can monitor single DNA molecules that are fluttering in the fluid flow with their one end anchored on a glass slide. Moreover, he can image single DNA molecules that are fully stretched and sit still on a glass surface. He captured dynamic cleaving process of the stretched DNA strands on real-time with a dose of restriction endonuclease solution. This technique could lead to a new approach for studying the kinetics of enzymatic hydrolysis.
Keywords
- Analytical chemistry
- Analytical instrumentation
- Analytical separation
- Fluorescence spectroscopy
- Genetic and epigenetic analysis
- Lab-on-a-chip
- Microfluidics
- Emulsion
- Point-of-care diagnosis
- Microfabrication
- Digital PCR
- Nucleic Acid